Development of nucleic acid amplification methods
Dr. Marras research has focused on developing fluorogenic probes for nucleic acid detection in nucleic acid amplification
assays, on solid surfaces, and in live and fixed cells. For the last two decades, he collaborated with Drs. Sanjay Tyagi and Fred Kramer on
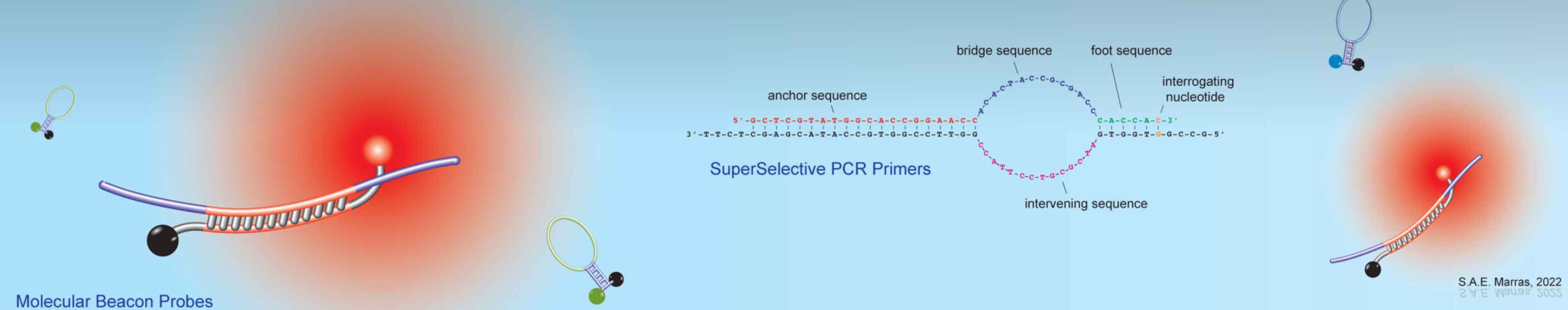
fluorescent nucleic acid hybridization probes, specializing in molecular beacon probes. He contributed to the molecular beacon
probe technology by establishing the thermodynamic parameters for their design, studying the mechanism of fluorescence
energy transfer between fluorescent labels and quenchers of fluorescence, and co-developing "color-coded," "sloppy," and
"wavelength-shifting" molecular beacon probes. Since 2006, he has been an independent faculty member at PHRI and have shared
laboratory space with Drs. Tyagi and Kramer. He collaborated with many different groups in academics and industry on designing,
optimizing, and troubleshooting real-time nucleic acid amplification assays - PCR and isothermal-based assays (NASBA, NEAR,
and LAMP). He co-invented SuperSelective primer technology, which enables the detection and quantification of somatic
mutations, whose presence relates to cancer diagnosis, prognosis, and therapy, in real-time multiplex PCR assays that are able to analyze
rare DNA fragments present in liquid biopsies. However, SuperSelective primer applications are not limited to cancer diagnosis, as
they can be used to identify and quantify any rare mutant DNA fragment in a large background of wild-type DNA fragments. He is a
co-inventor on fifteen issued US patents and their international counterparts, describing novel fluorescent nucleic acid hybridization
probes, hybridization strategies, and nucleic acid amplification PCR primers. He established a DNA Synthesis Core Facility at
PHRI. The facility includes a K&A H-4 oligonucleotide synthesizer, which simultaneously synthesizes four multi-labeled probes and/or
primers, two HPLC systems for probe purification, and a real-time thermal cycler for pilot PCR assay validation. The time from
design to validating probes and primers in a PCR assay is less than 36 hours. His expertise in the design and synthesis of
oligonucleotides, multi-labeled nucleic acid hybridization probes, fluorescence chemistry, and quantitative real-time nucleic acid
amplification assays is instrumental in establishing multiplex PCR assays for any genotype and pathogen identification assay.